Microbial Interaction

- Associate Professor
- WATANABE Daisuke

- Assistant Professor
- AKASAKA Naoki

- Labs HP
- https://bsw3.naist.jp/microbial_interaction/en/top/
Outline of Research and Education
How do microorganisms behave and interact to build complex ecosystems? We study yeasts and other unicellular organisms familiar to humans at the molecular, metabolic, cellular, and ecological levels to deepen understanding of diversity in the microscopic world. Our achievement will also contribute to modern biotechnology in food and health science (Fig. 1).
Major Research Topics
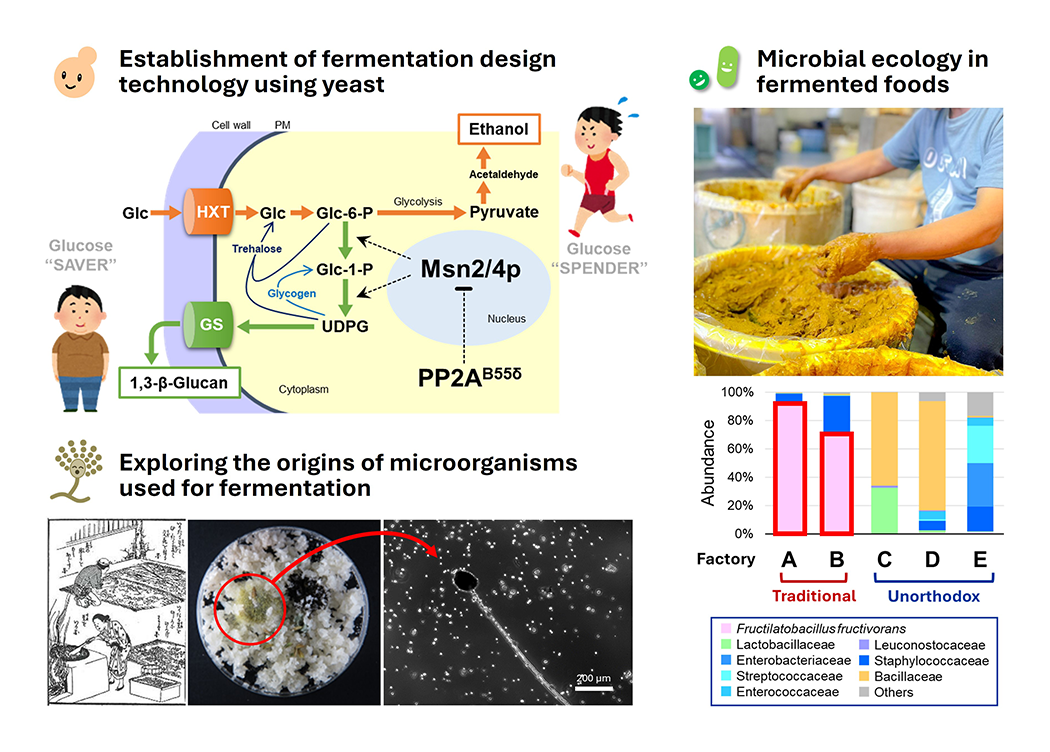
Microbial ecology in traditional food fermentation (Fig.2)
The rich flavors and health benefits of fermented foods have been attracting significant attention both domestically and internationally, leading to a growing demand. By deepening our understanding of fermented foods and the microorganisms that inhabit them—such as yeast, lactic acid bacteria, and koji fungi—we aim to pioneer the future of food for the next generation. The microscopic world, invisible to the human eye, remains full of mysteries about microbial behavior and interactions. In particular, we explore how the complex microbial ecosystems found in traditional fermented foods like sake, soy sauce, and preserved vegetables are structured and function. We also undertake innovative research focused on local fermented foods unique to Nara, a place once renowned as an ancient "capital of fermentation." Through these studies, we strive to elucidate the principles governing microbial ecosystem formation and environmental adaptation mechanisms while leveraging traditional knowledge. Our ultimate goal is to define the essence of fermentation.
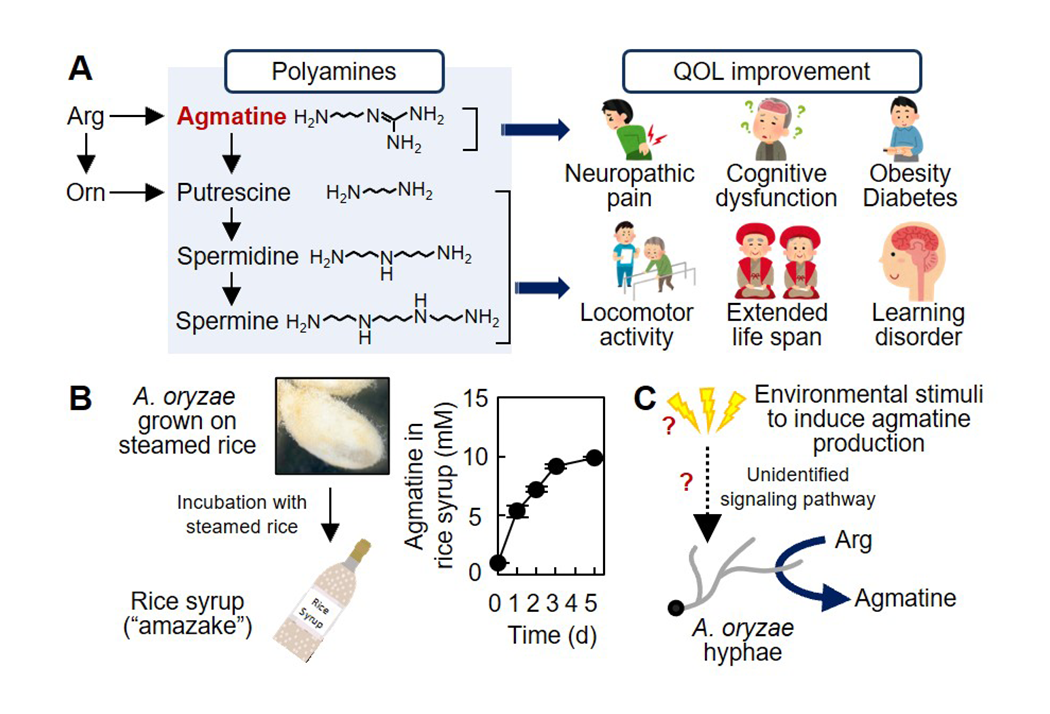
Health-promoting effect of fermented food ascribed to Aspergillus oryzae (Fig. 3)
A variety of traditional fermented foods have been produced since ancient times in Japan, and have been contributing to Japanese healthy life span. However, why the fermented foods are beneficial for our health remains scientifically well understood. Our major aim is to clarify the health-promoting effects of fermented foods from the view point of microbiology. The filamentous fungus A. oryzae is a key microbe in the production of various fermented foods as it produces hydrolyzing enzymes to degrades raw materials such as grains and soy bean. Our recent studies have revealed that A. oryzae produces significant amount of agmatine, a natural polyamine that improves the quality of life (QOL), highlighting the significance of A. oryzae not only in the source of hydrolyzing enzymes but also in the enhancement of our health. We are investigating the molecular mechanisms of the physiologically active polyamine production by A. oryzae. The resultant findings are expected applicable for the development of novel fermented foods and nutraceuticals with increased functionalities to promote healthy life span in aging/aged society.
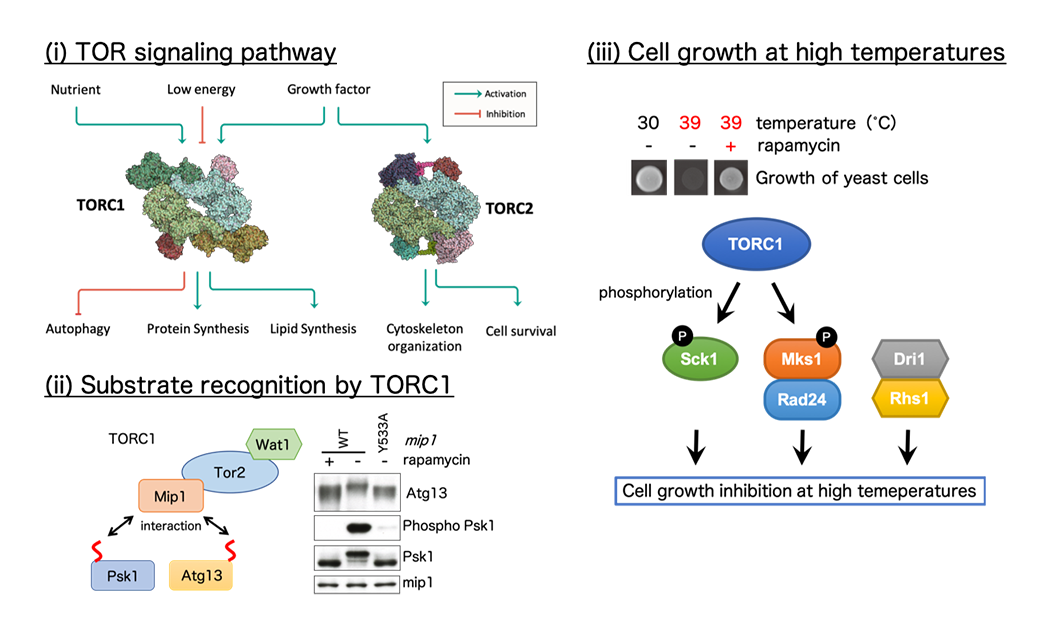
TOR (Target of Rapamycin) signaling pathway (Fig. 4)
The target of rapamycin (TOR) kinase forms two distinct multi-subunit complexes termed TORC1 and TORC2 to regulate cellular growth and proliferation in response to diverse stimuli, such as nutrients and growth factors. Deregulation of TOR signaling is often associated with human diseases, including cancers, diabetes, and neurodegenerative disorders, and therefore, comprehensive understanding of TOR pathway is critical to develop informed strategies to treat the diseases. We have established a model system in the fission yeast S. pombe to discover molecular mechanisms that control TORC1 and TORC2. We have recently discovered that TORC1, a well-established growth promoter, negatively regulates growth of fission yeast cells at high temperatures.
References
- Morozumi et al., FEBS Lett., 598, 2886-2896, 2024
- Murakami et al. Appl. Environ. Microbiol., 90, e0029424, 2024
- Morozumi et al., iScience, 27, 108777, 2024
- Watanabe et al., Int. J. Mol. Sci., 25, 304, 2024
- Tai et al., Mol. Cell Biol., 43, 675-692. 2023
- Watanabe et al., NPJ Sci. Food, 7, 37, 2023
- Watanabe and Hashimoto, Sci. Rep., 13, 9279, 2023
- Morozumi et al., J. Cell. Sci., 134, jcs258865, 2021
- Ishii et al., J. Bacteriol., 203, e0016221, 2021

 NAIST Edge BIO
NAIST Edge BIO