Environmental Microbiology

- Professor
- YOSHIDA Shosuke

- Assistant Professor
- Chek Min Fey

- Liu Yuting

- KOBAYASHI Kazuo

- Labs HP
- https://bsw3.naist.jp/ssk-yoshida/index-en.html
Outline of Research and Education
Human beings have placed a heavy burden on the environment through modern mass production/consumption of petrochemical products which are not circulable. Microbes live in all environments and are deeply involved in the global homeostasis. Recently, we have discovered a microbe that degrades a plastic which was thought not to be biodegraded. Why do microbes possess such unique abilities? How did they attain them? To answer these questions, we study microbial molecules and assemblies. We believe that our studies will lead to solutions for the sustainable development of society.
Major Research Topics
Elucidation of a bacterial PET metabolism
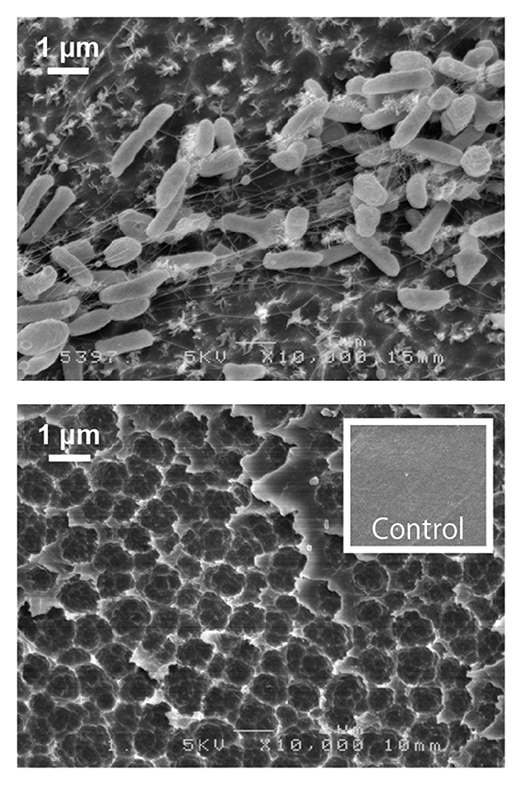
Poly(ethylene terephthalate) (PET) is a material used for plastic bottles and polyester fibers. A bacterium that we discovered named Ideonella sakaiensis can degrade and metabolize PET. The fact that this bacterium nutritionally utilizes PET has been revealed through discoveries such as unique PET hydrolyzing enzymes. By unraveling bio-information such as genomes and transcriptomes and using genetic and biochemical methods, we aim to fully understand the molecular mechanisms involved in PET degradation.
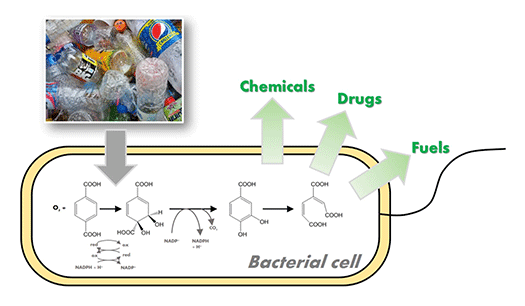
Fermentation of plastics
I. sakaiensis can eat PET. In other words, it has a metabolic system that can degrade and convert PET into energy and cellular components. We are attempting to breed the strains that produce high value compounds from waste PET products by modifying and/or enhancing their metabolism.
Visualizing microbiology
Microbial research has been focused on analysis of cells that can be observed with an optical microscope, or molecules that can be followed by their presence such as enzymatic reactions. However, in recent years, it has been found that many microbes secrete much smaller structures than their cells. To open this new microbial world, we are trying to clarify the functions of these nanostructures using electron and super-resolution microscopes.
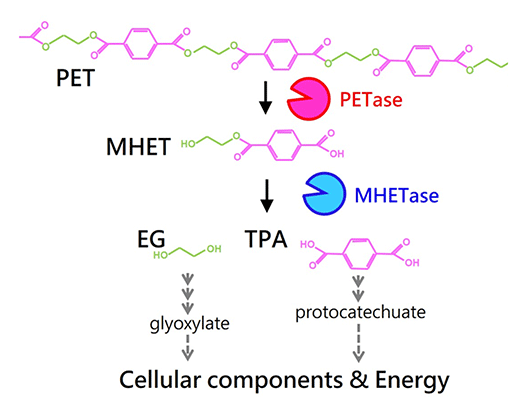
Two unique enzymes, PETase and MHETase, are able to efficiently convert PET into its monomers. These monomers are further metabolized by this strain.
References
- Fujiwara et al., Sci. Rep., 11, 2021
- Hachisuka et al., Appl. Environ. Microbiol., 87, 2021
- Yoshida S. et al., Methods Enzymol., 648, 2021
- Hiraga K. et al., EMBO Rep. 21, e49826, 2020
- Taniguchi, I.*, Yoshida, S.* (*equally contributed), et al., ACS Catal. 9, 4089-4105, 2019
- Yoshida S. et al., Science 351, 1196-1199, 2016
- Tanasupawat S. et al., Int. J. Syst. Evol. Microbiol. 66, 2813-2818, 2016
- Yoshida S. et al., Biosci. Biotechnol. Biochem. 79, 1965-1971, 2015
- Yoshida S. et al., Biochemistry 50, 3369-3375, 2011
- Nishitani Y.*, Yoshida S.* (*equally contributed) et al., J. Biol. Chem. 285, 39339-39347, 2010
- Yoshida S. et al., J. Bacteriol. 192, 5424-5436, 2010
- Yoshida S. et al., J. Bacteriol. 192, 483-493, 2010
- Yoshida S. et al., Appl. Environ. Microbiol. 73, 6254-6261, 2007

 NAIST Edge BIO
NAIST Edge BIO