Bioengineering
- Professor
- KATO Ko

- Assistant Professor
- WAKABAYASHI Tomomi

- KATO Takehide

- TAMURA Taizo

- Labs HP
- https://bsw3.naist.jp/ko-kato/e-index.html
Outline of Research and Education
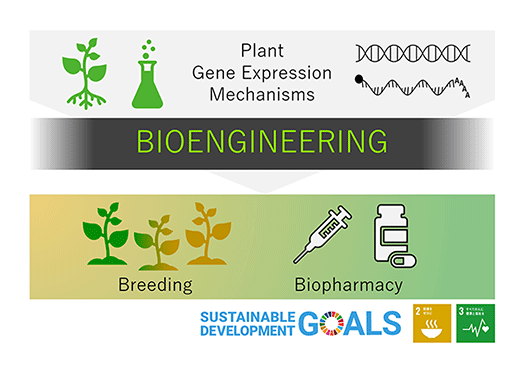
To contribute to society through biotechnology, we are developing basic technologies for high production of useful proteins in plants and are revealing mechanisms of plant phenotype controls, by understanding the mechanism of gene expression regulation (Fig. 1).
We provide guidance so that students can understand the research logically and broaden and develop the base of knowledge. In addition to regular laboratory meetings, we invite researchers and practitioners from industry to introduce the knowledge necessary for research and development in companies. Through these guidances, we aim to develop human resources who can play an active role in a low-growth, global society.
Major Research Topics
Isolation and improvement of elements involved in high expression of transgene
The production of useful proteins such as medical vaccines and growth factors using plants as hosts is attracting a lot of attention. To increase protein productivity, it is essential to elucidate the sequence characteristics that are involved in determining efficiency and to optimize the nucleotide sequence of the introduced gene (Fig. 2). Our laboratory uses next-generation sequencers, machine learning, and optimization algorithms to analyze core promoters involved in transcription, as well as 5'UTR and 3'UTR sequences that are involved in mRNA stability and translation efficiency. We are working to develop systems that design completely new arrays that exceed existing efficiency. The results have been provided to multiple companies, and some of the results have already been put into practical use.
Elucidation of the mechanisms of phenotypic control and adaptive evolution by gene expressions
In this research, we are focusing on traits related to the plant fitness, such as flowering time. Based on the whole-genome sequences and gene expressions, we are trying to detect the responsible genes for the traits (Fig. 3) and how the genes are related to the phenotype. We aim to understand the mechanisms of reproductive success and adaptive evolution pf wild plants.
Elucidation of the environmental adaptation mechanism of plants through gene expression
This research focuses on the ability of plants to skillfully withstand environmental stress conditions and the mechanisms by which they flexibly adapt to environmental changes (Fig. 4). Using strains with different environmental adaptability, we search for responsible genes through genome analysis. We aim to elucidate response mechanisms to environmental changes through high-resolution gene expression analysis.
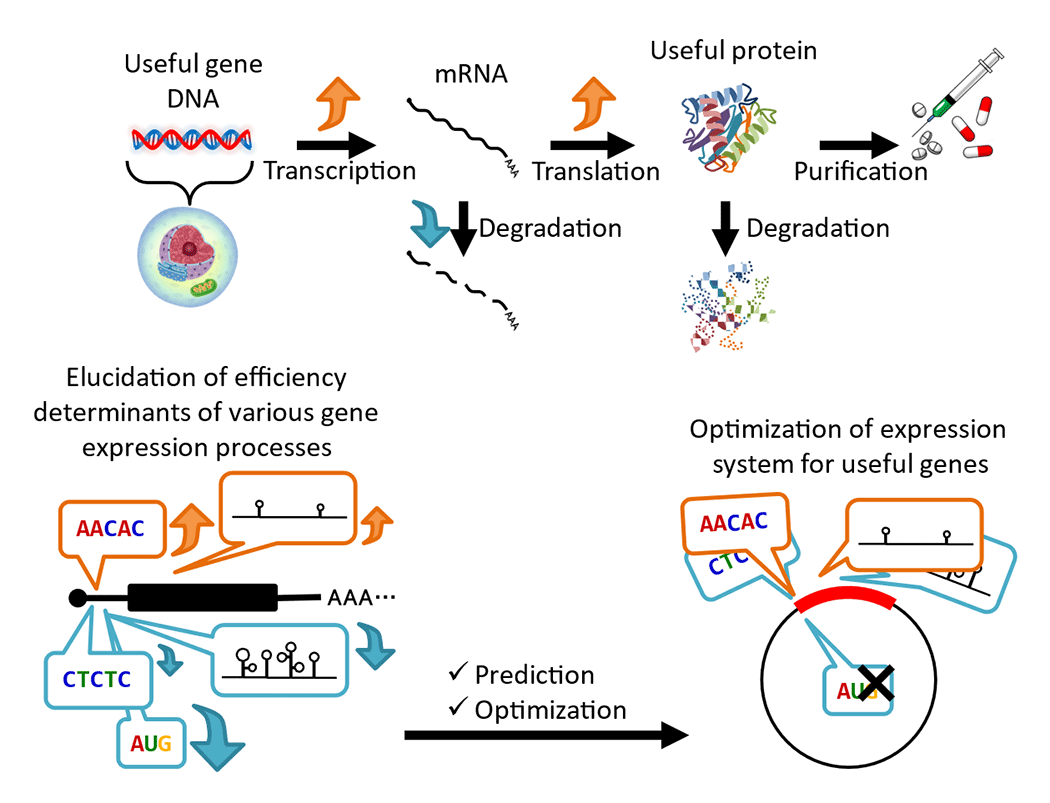
In order to optimize each step of gene expression, we are analyzing each regulatory process in detail.
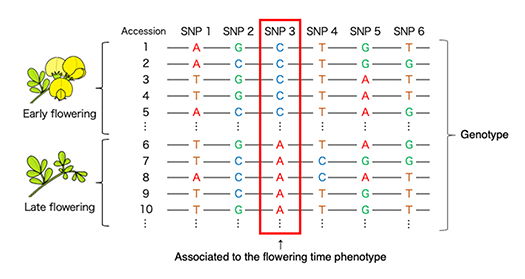
Analyzing genome-wide genotype data and phenotype data of accessions enables estimation of genome regions associated to target traits.
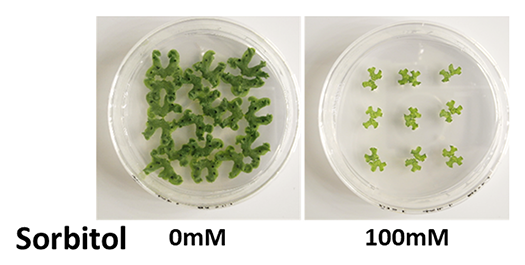
3 weeks in medium with/without sorbitol. Growth is inhibited in this strain due to the high osmotic pressure of the medium.
References
- Wakabayashi et al., Plant Biotechnol., in press
- Hattori et al., LSSR., in press
- Ueno D. et al., J. Biosci. Bioeng., 143, 450-461, 2022
- Ueno D. et al., Plant Sci., 318, 111241, 2022
- Ueno D. et al., BMC Bioinform., 22, 380, 2021
- Matsui T. et al., Plant Biotechnol., 38, 239-246, 2021
- Ueno D. et al., Plant Cell Physiol., 62, 143-155, 2021
- Ueno D. et al., Plant Cell Physiol., 61, 53-63, 2020
- Shah N. et al., Nat. Commun., 11, 253, 2020

 NAIST Edge BIO
NAIST Edge BIO