Systems Neurobiology and Medicine
- Professor
- INAGAKI Naoyuki

- Assistant Professor
- MINEGISHI Takunori

- Labs HP
- https://bsw3.naist.jp/inagaki/english/
Outline of Research and Education
Neurons extend axons, and form elaborate networks in our brain; all the brain activities depend on neuronal networks. To establish proper neuronal networks, axons decide their migratory route in response to gradients of chemical signals in the brain with extraordinary sensitivity. But, little is known about how axons are able to decide their migratory routes by reading gradients of chemical signals and by translating them into directional driving force.
In addition to axons, various cells migrate within our body, thereby playing key roles in organ formation, immune responses, wound healing and regeneration. Disruption of axon guidance and cell migration is implicated in diseases, including birth abnormality, neuronal disabilities, immune disorders and cancer metastasis.
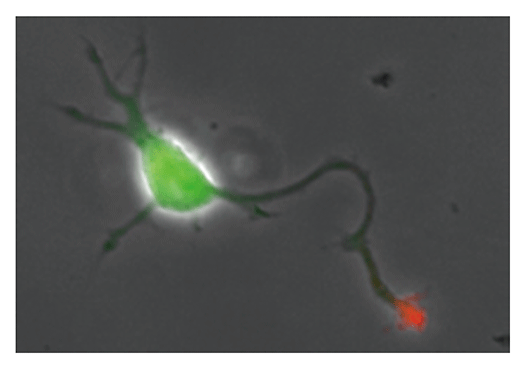
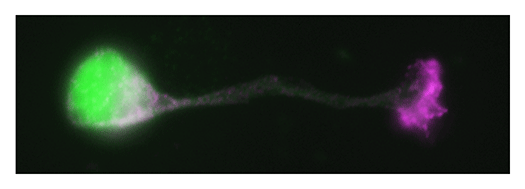
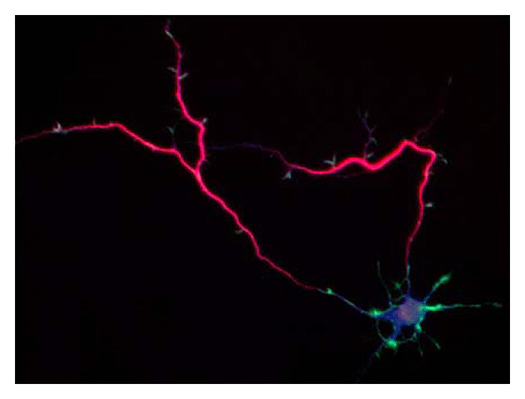
Our laboratory focuses on the proteins, Shootin1a (Fig. 1) , Shootin1b (Fig. 2) and Singar (Fig. 3), which we identified by proteome analyses, as well as their interacting proteins, Cortactin, L1-CAM and Rab33. We are analyzing the molecular mechanisms for axon formation, axon guidance, cell migration, and synaptic plasticity. We also analyze actin waves, which is a new type of intracellular protein transport system for cell morphogenesis and cell migration (Fig. 4) .
We expect that our studies will help us to understand the mechanisms underlying neuronal morphogenesis and neuronal network formation, as well as the mechanisms underlying diseases including birth abnormality (Fig. 6), neuronal disabilities, neuropsychiatric disorders and immune disorders, giving us a new window into therapeutic strategies for nerve injury, Alzheimer’s disease, neuropsychiatric disorders and cancer metastasis.
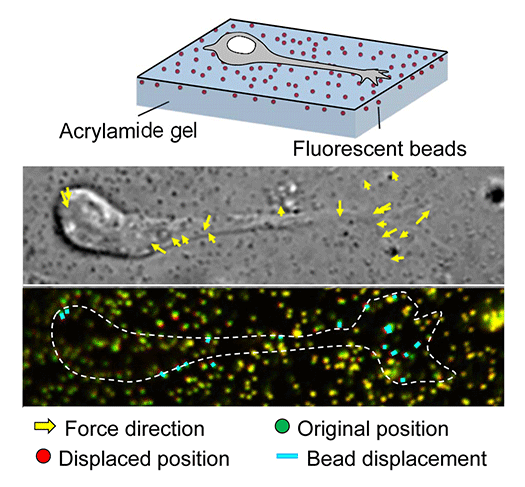
In our laboratory, we use standard biological methods (biochemistry, molecular biology, cell biology and developmental biology), as well as cutting-edge technologies including single molecule imaging, systems biology (Fig. 5) , mechanobiology (Fig. 7), and laser manipulation. Yet, we do not ask previous experiences to join our laboratory. We will educate these methods from basics step-by-step: starting research in our laboratory is not difficult.
Major Research Topics
Axon/dendrite formation and neuronal polarization
The basic function of neurons is to receive signals, integrate them and transmit them to the next neuron. Thus, neurons can function as semiconductors in the brain. Neuronal polarity plays an essential role for this function. Namely, most of neurons form a single axon and multiple dendrites. Dendrites receive signals from outside, while an axon output signals. Then a signal flow arises from dendrites via cell body to the axon. How neurons acquire neuronal polarity? Our laboratory investigates the mechanism for axon/dendrite formation and neuronal polarization, focusing on Shootin1a (Fig. 1) , Singar (Fig. 3) and Rab33.
Mechanical forces for axon guidance and cell migration
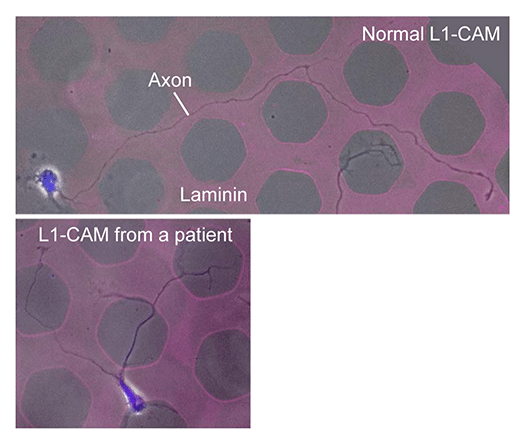
Animals move and change their shape, using force generated by muscles. How cells do so? Actin polymerizes near the leading edges of neurite tips and migrating cells. We reported that shootin1a interacts with both the polymerizing actin filaments and the cell adhesion molecule L1-CAM at neurite tips, thereby generating traction force for neurite outgrowth. We are currently analyzing molecular mechanisms for generation and regulation of mechanical forces (Fig. 7) for axon guidance and neuronal migration. We also reported a new type of axon guidance mechanism (Grip & Slip mechanism) induced by the extracellular matrix protein Laminin.
Actin waves: a new mechanism for cellular protein transport
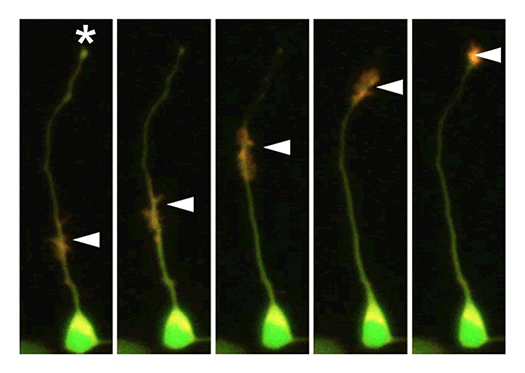
Actin filaments and associated proteins undergo wave-like movement in various cell types (Fig. 4). We reported that axonal actin waves migrate by means of directional assembly and disassembly of membrane-anchored actin filaments. Thus, the actin wave in axons represents a new type of machinery that translocates actin and associated proteins to the cell edge. The involvement of actin waves in axon outgrowth was also demonstrated by laser-manipulation of the adhesion substrate. We are currently analyzing roles of actin waves in cell polarity formation and migration, introducing single molecule imaging and mathematical modeling (Fig. 5).
Research in medicine: brain diseases and cancer metastasis
We expect that our studies will help us to understand the mechanisms underlying diseases including birth abnormality, neuropsychiatric disorders, and immune disorders, giving us a new window into therapeutic strategies for nerve injury, Alzheimer’s disease, neuropsychiatric disorders, and cancer metastasis. We recently reported “Grip & Slip mechanism” for axon guidance; this mechanism is disrupted in a human patient of L1-CAM/CRASH syndrome, suffering corpus callosum agenesis and corticospinal tract hypoplasia. We are also currently analyzing the molecular mechanisms for immune cell migration and tumor cell metastasis.
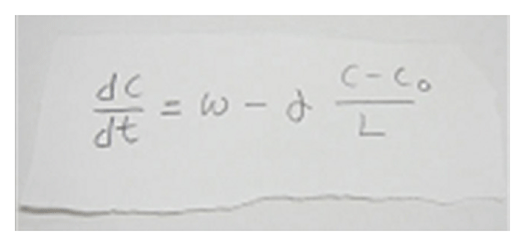
References
- Baba, K. et al., Adv. Sci., e02074, 2025
- Minegishi, T. et al., EMBO J, 44, 767-780, 2025
- *Qiu, Z. et al, Front Mol Neurosci, 17, 1307755, 2024
- Kastian, R.F. et al., J Biol Chem, 299, 104687, 2023
- Minegishi, T. et al., Sem. Cell Dev. Biol. 140, 82-89, 2023
- *Kastian, R.F. et al., Cell Rep, 35, 109130, 2021
- Abe, K. et al., Biophys J, 120, 3566-3576, 2021
- Minegishi, T. and Inagaki, N., Front Cell Dev Biol, 8, 863, 2020
- *Huang L. et al., Sci. Rep. 9:1799, 2019
- Urasaki A. et al., Sci. Rep., 9, 12156, 2019
- *Minegishi T. et al., Cell Rep., 25, 624-639, 2018
- *Baba K. et al., eLife, 7, e34593, 2018
- *Abe et al., PNAS, 115, 2764-2769, 2018
- Inagaki N. and Katsuno H., Trends Cell Biol., 27, 515-526, 2017
- *Higashiguchi et al., Cell Tissue Res., 366, 75-879, 2016
- *Katsuno H. et al., Cell Rep., 12, 648-660, 2015
- *Kubo Y. et al., J. Cell Biol., 210, 663-676, 2015
- Toriyama M. et al., Curr. Biol. , 23,529-534, 2013
- *Nakazawa H. et al., J. Neurosci., 32, 12712-12725, 2012
- Inagaki N. et al., Dev. Neurobiol., 71, 584-593, 2011
- Toriyama M. et al., Mol. Syst. Biol., 6, 394, 2010
- Shimada T. et al., J. Cell Biol., 181, 817-829, 2008
- *Mori T. et al., J. Biol. Chem., 282, 19884-19893, 2007
- *Toriyama M. et al., J. Cell Biol., 175, 147-157, 2006
- Inagaki and Katsuta, Curr. Proteomics 1, 35-39, 2004
- **Nomura E. et al., J. Mass Spectrometry, 39, 666-672, 2004
- **Oguri T. et al., Proteomics, 2, 666-672, 2002
- Fukata Y. et al., Nature Cell Biol., 4, 583-591, 2002
- Inagaki N. et al., Nature Neurosci., 4, 872-873, 2001
- Inagaki N. et al., J. Biol. Chem., 275, 27165-27171, 2000
(*Papers by doctor course students; **papers by master course students)

 NAIST Edge BIO
NAIST Edge BIO