Data-driven Biology
- Professor
- SAKUMURA Yuichi

- Assistant Professor
- KOKAJI Toshiya

- SUZUKI Kenta

- Labs HP
- https://sites.google.com/sakulab.org/w3en
Outline of Research and Education
Our laboratory aims to elucidate the underlying principles of phenomena spanning from the molecular level to human population dynamics by mathematically analyzing experimental and observational data. By utilizing information and mathematical analysis, we quantitatively link phenomena across different scales and seek to understand biological functions as interactions among various physical quantities. Through these studies, we develop data analysis techniques and cultivate independent thinking skills.
Major Research Topics
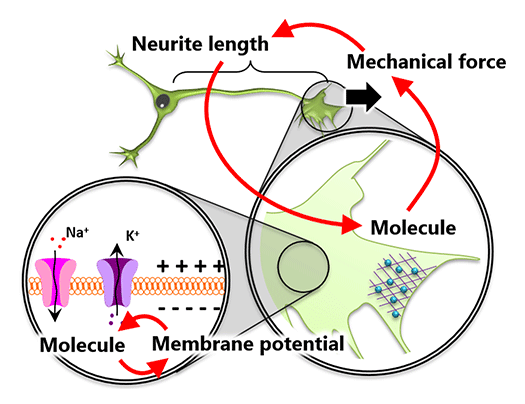
Systems Biology of Cell Deformation and Migration (From molecules to cells) (Fig.1)
The action potential of neurons represents one aspect of a system composed of membrane proteins and membrane potential. Similarly, cell deformation and migration can be understood as a system where molecules, forces, and morphology interact (Figure 1). To elucidate these systems governed by signals of different physical scales, it is essential to derive mathematical relationships among the data.
- Elucidation of the system regulating cytoskeletal formation, force control, and cell deformation
- Estimation of cellular traction forces using machine learning
- Quantitative modeling of the viscoelastic properties of cellschange.
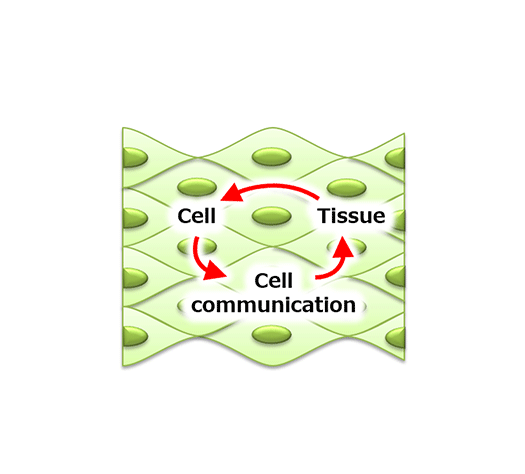
Systems Biology of Tissue Formation (From cells and tissues) (Fig. 2)
Biological tissues function as integrated systems of cells and tissues (Figure 2). Through mathematical analysis of experimental data, we aim to elucidate the principles by which cellular communities generate resilient, rather than robust, tissue functions.
- Cellular coordination during vertebrate development
- Angiogenesis based on the principles of cell migration
Medical Systems Biology (From molecules and tissues to human diseases)
We aim to elucidate the systems underlying human diseases through mathematical analysis of medical data, spanning molecular and tissue levels. Our research involves disease diagnosis and prediction using breath and blood components, as well as tissue imaging. Additionally, we apply machine learning techniques to analyze large-scale medical claims data, enabling disease risk estimation and the extraction of disease-related factors.
- Disease diagnosis and prediction using breath and blood components
- Disease risk estimation through large-scale medical data and machine learning
- Disease prediction using MRI imaging
- Extraction of disease-associated genes
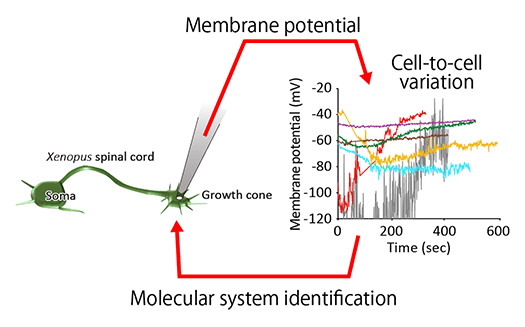
Preprocessing and Machine Learning Applications for Biological Data Analysis (Fig. 3)
Extracting common insights from complex biological data at the cellular and organismal levels is no longer feasible solely through researchers' intuition. Our laboratory applies data preprocessing techniques and machine learning to uncover underlying patterns and identify key factors driving biological phenomena.
- Trans-omics analysis
- Estimation of mechanical forces generated by cells using machine learning
- Inference of gene regulatory networks involved in tissue formation
- Data-driven reproductive control in model organisms
References
- Suzuki et al, Sci Rep, doi:10.1038/s41598-025-91034-8, 2025
- Ohkubo et al, Sci Rep, doi: 10.1038/s41598-024-76029-1, 2024
- Fujikawa et al, Biophys J, doi: 10.1016/j.bpj.2023.10.032, 2023
- Ohkubo et al, Sci Rep, doi: 10.1038/s41598-023-40469-y, 2023
- Itoh et al, Sens Actuators B Chem, doi: 10.1016/j.snb.2023.133803, 2023
- Kunida et al, Cell Rep, doi: 10.1016/j.celrep.2023.112071, 2023
- Minegishi et al, J Vis Exp, doi: 10.3791/63227, 2021
- Wong et al, Plant Physiol, doi:10.1104/pp.19.00811, 2019

 NAIST Edge BIO
NAIST Edge BIO