Research outcomes
Timing is everything: New insights into floral development unveil nature's perfect clock
Arabidopsis floral meristem termination is controlled by an epigenetic biotimer that determines the concentration of histone methylation regulators in a cell-cycle dependent manner
The intricate process of flower development has long fascinated scientists seeking to unravel the mysteries behind nature's precision timing. In a study published in the journal Plant Cell, a research team led by Nara Institute of Science and Technology (NAIST), Japan has shed light on the inner workings of floral meristem termination and stamen development, uncovering a unique mechanism driven by the interplay of genetic and epigenetic factors.
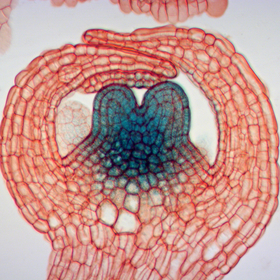
Flowers owe their intricate structures to delicate stem cell differentiation, a process by which founder cells develop into specialized cells in floral meristems. However, the precise moment at which stem cells cease self-renewal and transform into their form remains largely unknown. Driven by a desire to decipher this critical temporal transition, the researchers turned their attention to AGAMOUS (AG), a pivotal MADS domain transcription factor governing floral meristem termination.
Through meticulous investigations in the model plant Arabidopsis thaliana , the team discovered that AG serves as a master conductor, orchestrating gene expression through a process known as cell cycle-coupled H3K27me3 dilution. This remarkable phenomenon involves the dilution of a histone modification called H3K27me3 along specific gene sequences, effectively kickstarting gene activation. The scientists identified several key genes directly regulated by AG at various time points of this cycle.
The study revealed a genetic network tightly controlled by AG, with genes such as KNUCKLES (KNU ), AT HOOK MOTIF NUCLEAR LOCALIZED PROTEIN18 (AHL18 ), and PLATZ10 emerging as critical players. "By unraveling the inner workings of this regulatory circuit, we gained unprecedented insight into the intricate timing mechanisms that drive proper floral meristem termination and stamen development," says first author Margaret Anne Pelayo.
To unlock the secrets of this remarkable system, the researchers devised a mathematical model capable of predicting gene expression timing with astonishing accuracy. By modifying the length of H3K27me3-marked regions within the genes, they successfully demonstrated that gene activation could be delayed or reduced, confirming the influence of this epigenetic timer. The team's findings offer a novel perspective on how nature controls the gene expression during flower development.
Furthermore, their study identified AHL18 as a stamen-specific gene with a profound impact on stamen growth and development. Misexpression of AHL18 led to intriguing developmental defects, highlighting the gene's vital role in ensuring proper stamen elongation and maturation. Additionally, the team discovered that AHL18 selectively binds to genes crucial for stamen growth, uncovering a new layer of regulatory complexity in flower development.
Nobutoshi Yamaguchi, senior author of the study, opined that this research not only deepens our understanding of the mechanisms underpinning floral development but also presents a potential tool for fine tuning gene expression patterns. Manipulating the delicate balance of epigenetic modifications opens up exciting possibilities for controlling plant reproduction in a flexible and reversible manner, ultimately benefiting our food supply and agricultural practices.
This study paves the way for further exploration of epigenetic approaches to regulate gene expression with exquisite precision. By unraveling nature's perfect clock, scientists may one day unlock new strategies for enhancing crop productivity and bolstering plant resilience, and contribute to food security despite environmental challenges.
###
Resource
・Title: AGAMOUS regulates various target genes via cell cycle-coupled H3K27me3 dilution in floral meristems and stamens
・Authors: Margaret Anne Pelayo, Fumi Morishita, Haruka Sawada, Kasumi Matsushita, Hideaki Iimura, Zemiao He, Liang Sheng Looi, Naoya Katagiri, Asumi Nagamori, Takamasa Suzuki, Marek Širl, Aleš Soukup, Akiko Satake, Toshiro Ito, Nobutoshi Yamaguchi
・Journal: Plant Cell
・DOI: 10.1093/plcell/koad123
・Information about the Plant Stem Cell Regulation and Floral Patterning Laboratory can be found at the following website: https://bsw3.naist.jp/eng/courses/courses112.html
( May 31, 2023 )
